DDI is a flexible standard and different users are at will to implement the standard in slightly different ways. This is a strength, but when moving between different implementations some adjustments need to be made.
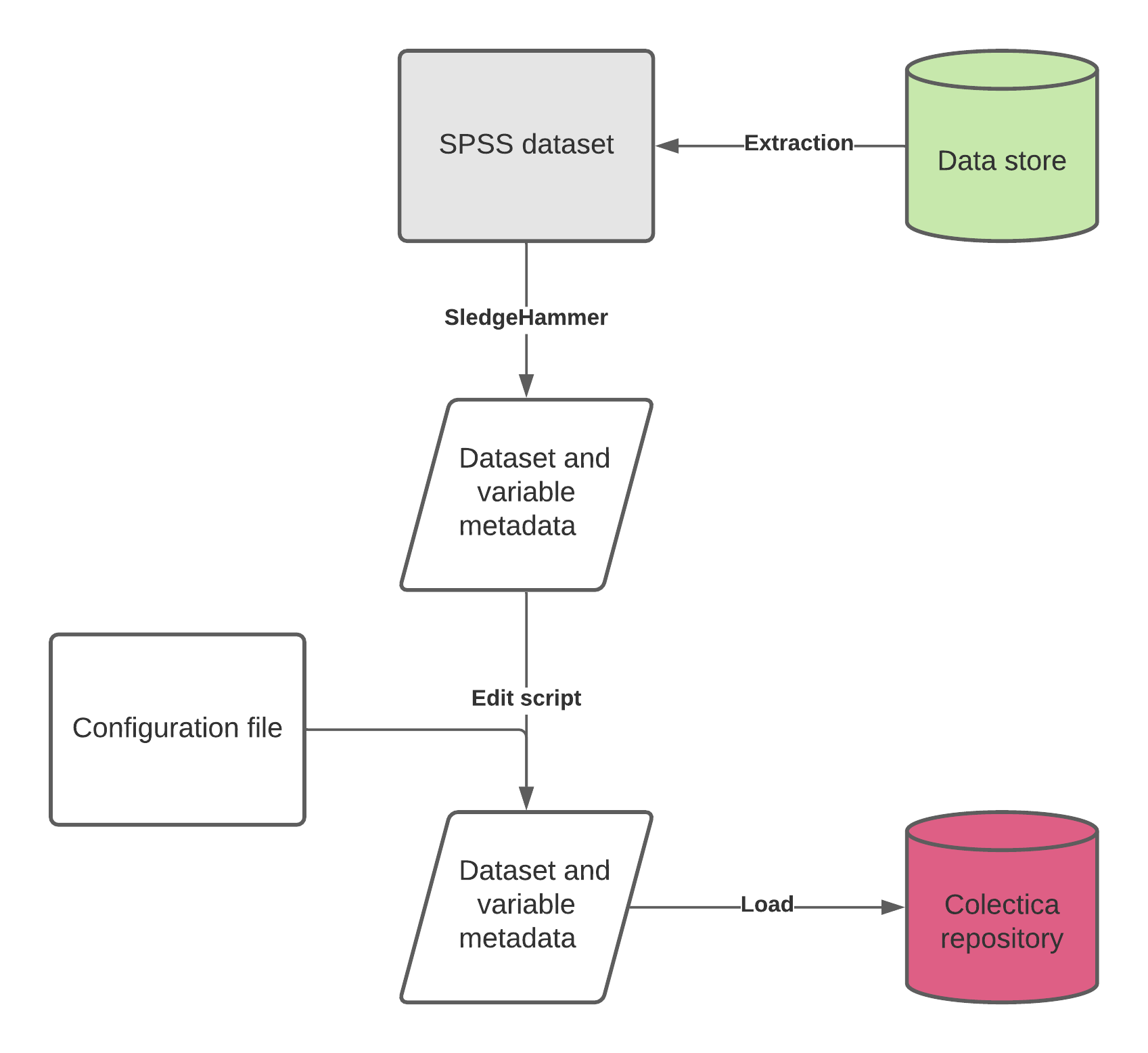
Data files should meet certain criteria, this generates a standard SledgeHammer output, which is then lightly edited to provide a consistent structure for ingest into Colectica Repository.
The following provides a detailed overview of the processes involved. Please see the step by step guide for the simple steps.
Variable metadata workflow
Data Files
Studies should be encouraged to generate data files that are of the standards the UK Data Service
- use meaningful and self-explanatory variable names, codes and abbreviations
- variable and value labels must be clear and consistent, avoiding truncation of variable and value labels
- non-compliant characters, such as &, @ and <>, should be removed
- ensure no repetition of variables, especially redundancy in derived variables
- ensure consistent treatment and labelling of missing values
- extraneous information such Document entries should be stripped out
The workflow expects a simple mapping of dataset to questionnaire. The naming of the datafile should be consistent with that of the questionnaire it is collected from.
Data files should ideally be in SPSS format. Some guidance on SPSS file preparation is given below.
SPSS File Preparation
SPSS will hold lots of hidden information, Sledgehammer will try to use this and can lead to issues when outputting the DDI-L XML. We would recommend using something like this to get rid of this extraneous information This replaces a file label (often the location of the original file) with the bundle name, and to drop any document(s):
get file="G:\DB\closer_data\bcs70\bcs_1975\bcs_1975_masc.sav". FILE LABEL "bcs_75_msc". DROP DOCUMENT. EXECUTE. sysfile info file="G:\DB\closer_data\bcs70\bcs_1975\bcs_1975_masc.sav". save outfile="G:\DB\closer_data\bcs70\bcs_1975\bcs_1975_masc.sav".
Use of SledgeHammer
SledgeHammer is a product released by Metadata Technology North America (MTNA) and allows the extraction of metadata from a wide range of data formats. Although it can be run interactively, the project uses batch files to allow a consistent generation of output.
The project uses a restricted set of these commands:
| Command | Examples | Explanation |
|---|---|---|
| -ag | uk.cls.mcs, uk.alspac | Agency |
| -rename | alspac_00_ayc nshd_46_tcs | This should be the name of the bundle with which the data is associated |
| -ddi | Always 3.2-RP | DDI Version - this should not be changed |
| -ddipd | Always proprietary | Only output proprietary format not ascii |
| -har | “No options” | This creates unified codelists i.e. single Yes/No |
| -ddilang | Always en-GB | This should stay as en-GB as this is the default setting we will be using |
| -ddiref | Always URN | Internal ddi URN definition |
| -ddiurn | Always canonical | Canonical - this should not be changed |
| -pretty | “No options” | This is so it looks half decent if you look at by hand |
| -stats | min, max, valid, invalid | Description of statistics generated per variable Optional: stddev and freq |
| -opt | Always full | Optimised output |
| -scan | “No options” | Outputs metadata or entire full and includes no of cases and variables |
| ../bcs70/bcs_1970.sav | Name and path of input data file This is always the last line |
Batch File
Each dataset should have a batch or command file which calls the sledgehammer-cl.bat file, and lists the options above An example is:
sledgehammer-cl.bat" ^ -ag uk.cls.bcs70 ^ -rename bcs_75_mcs ^ -ddi 3.2-RP ^ -ddipd proprietary ^ -har ^ -ddilang en-GB ^ -ddiref urn ^ -ddiurn canonical ^ -pretty ^ -opt full ^ -scan ^ -stats max,min,mean,mode,valid,invalid,freq,stdev ^ ../bcs70/bcs_1975/bcs_1975_masc.sav
Metadata Edits (DDI flavour)
For display purposes and for ease of navigation and ingest, a consistent set of names should be applied to the output from SledgeHammer prior to ingest through a series of edit scripts. These are written in python, and if they cannot be run at the study, can be run at CLOSER prior to ingest.
| Edit script | Explanation |
|---|---|
| fandr.py | Insert <r:String> where absent from output |
| fandr2.py | Names the DDI Instance |
| fandr3.py | Names the Physical Instance |
| fandr4.py | Names the Logical Product |
| fandr5.py | Names the Code List scheme |
| fandr6.py | Names the Data Product Name |
| fandr7.py | Add Dataset URI and whether public |
| fandr8.py | Adds Title and Alternate Title to DDI Instance |
| fandr9.py | Corrects Valid to be ValidCases |
| fandr10.py | Corrects Invalid to be InvalidCases |
| fandr11.py | Adds naming to DataRelationship |
The scripts uses a tab delimited file called rename_list.txt which includes the following:
Short Name - is the name of the metadata bundle with which the dataset of associated with
Long Name - the name you want to display as a human readable description
DOI - if available, this allows the user to navigate to the DOI and relevant citation and is provided for the user. If this is not available a website address of where the data can be accessed can be used instead.
Public - 1 is to be used when an website address or DOI has been provided.
An example of this file is shown below.
| Short name | Long Name | DOI | Public |
|---|---|---|---|
| mcs_03_na | MCS2 Neighbourhood Assessment | http://dx.doi.org/10.5255/UKDA-SN-5350-3 | 1 |
| mcs4_teacher | MCS4 Teacher Survey | http://dx.doi.org/10.5255/UKDA-SN-6848-1 | 1 |
| mcs5_sc | MCS5 Child Paper Self-Completion | http://dx.doi.org/10.5255/UKDA-SN-7464-2 | 1 |
| mcs5_teacher | MCS5 Teacher Survey | http://dx.doi.org/10.5255/UKDA-SN-7464-2 | 1 |
| ncds8_sc | NCDS8 Paper Self-Completion | http://dx.doi.org/10.5255/UKDA-SN-6137-2 | 1 |
| pms | Perinatal Mortality Study | http://dx.doi.org/10.5255/UKDA-SN-5565-2 | 1 |
Control File
A control file can be used to batch up the batch files and then run the edits across all the files:
call pms.bat call ncds8_sc.bat call mcs_03_na.bat call bcs_1970.bat call mcs4_teacher.bat call mcs5_teacher.bat call mcs5_sc.bat call bcs_75_mcs.bat python g://db//bin//fandr.py python g://db//bin//fandr2.py python g://db//bin//fandr3.py python g://db//bin//fandr4.py python g://db//bin//fandr5.py python g://db//bin//fandr6.py python g://db//bin//fandr7.py python g://db//bin//fandr8.py python g://db//bin//fandr9.py python g://db//bin//fandr10.py
Outputs
For each dataset a DDI 3.2 file called [shortname].ddi32.rp.xml will be generated.
Checking
If the edits are run, the file can be imported into Colectica Designer to check that it is well formed. Alternatively you can check that DDI-Flavour has been run by checking the XML has the new title at the beginning of the file.